PhUSE US Connect-2021
June 2021
Integrating Clinical data for Translational Research
During development of biological or biomarker-based therapies, extracting insights from study data to assess efficacy for stratified cohort is a challenge because of the disparate nature of the data sources. The challenge is to use smart data transformation and machine learning algorithms to normalize and harmonize this disparate data to a single Universal Data Model (UDM), that supports any further analysis, visualization, or pattern recognition.
UDM can also enable integration of translational data such as biomarkers and assay data, with longitudinal subject data in legacy studies. Normalized and harmonized data in UDM is ready to be analyzed to gain insights on mechanisms, to perform cross-study analysis, and derive patient cohorts. In translational and precision medicine, these datasets can also be used as training sets for the machine learning algorithms designed to predict the outcomes of new drugs.
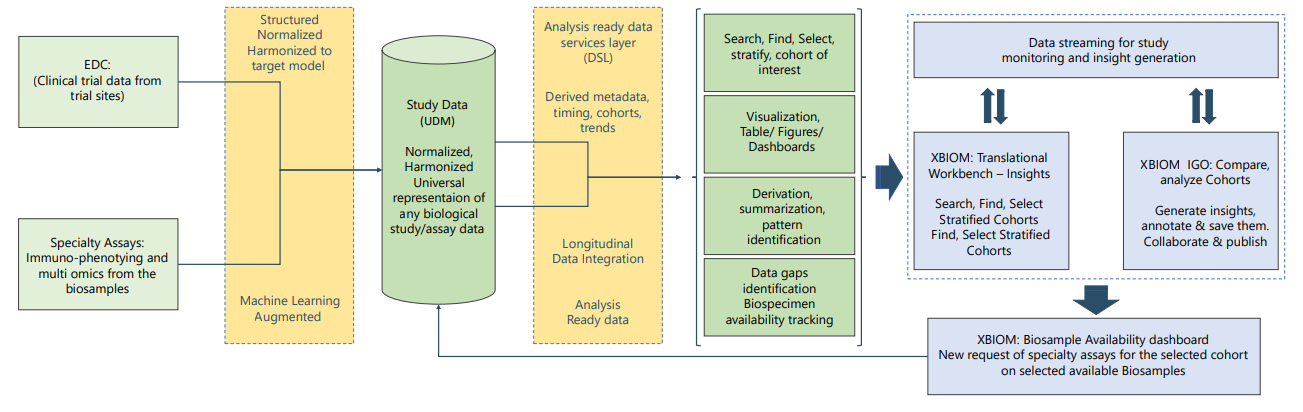
Data transformation for normalization and harmonization
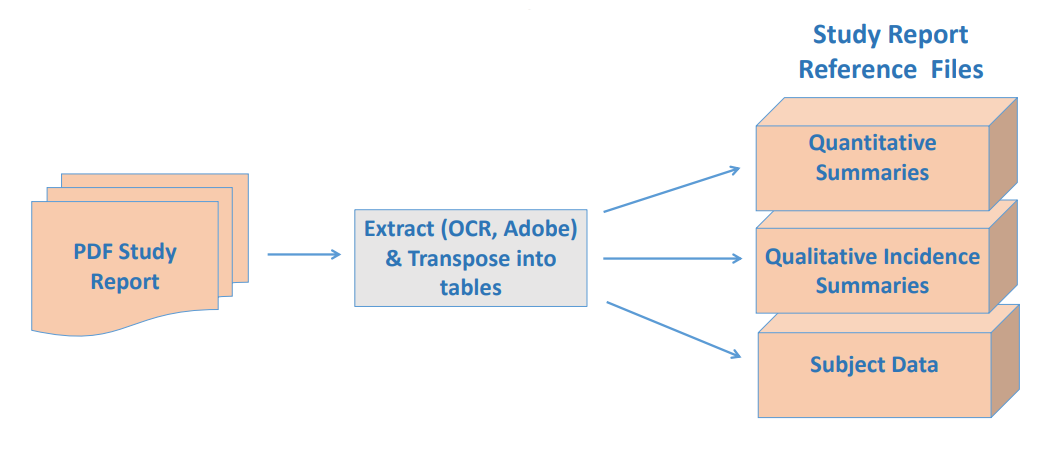
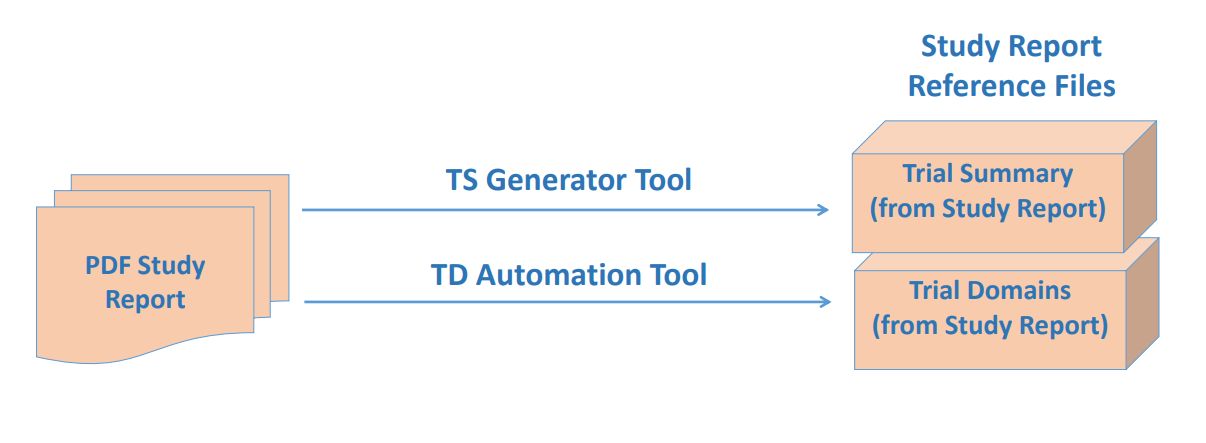
Data generated over a period in immunological studies/ cell/ gene therapy are very large and are often stored in many places. To make it available for insight generation at the right time and right place will require tools to standardize and integrate these data for their respective clinical data.
- Automation and recommendation engine using Machine Learning algorithms working on the principle of supervised learning will both speed up and standardize and harmonize the data to a chosen libraries/ontologies.
- Validation of the data for transformation and consistency.
- Indexed and stored in a customizable Universal Data Model.
- Metadata capturing at various levels of the data and making it available for search and selection of cohort.
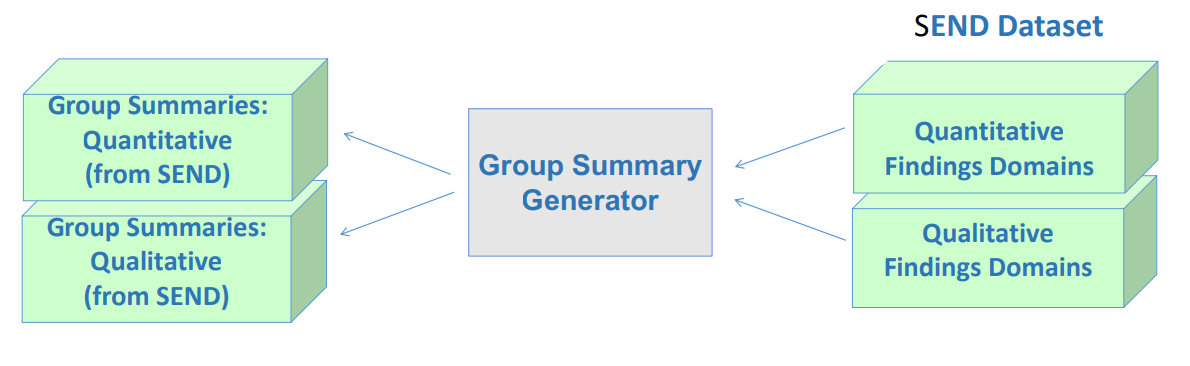
Derive metadata for analysis, insights, and cohort selection
- Statistical tools for visualization and analysis
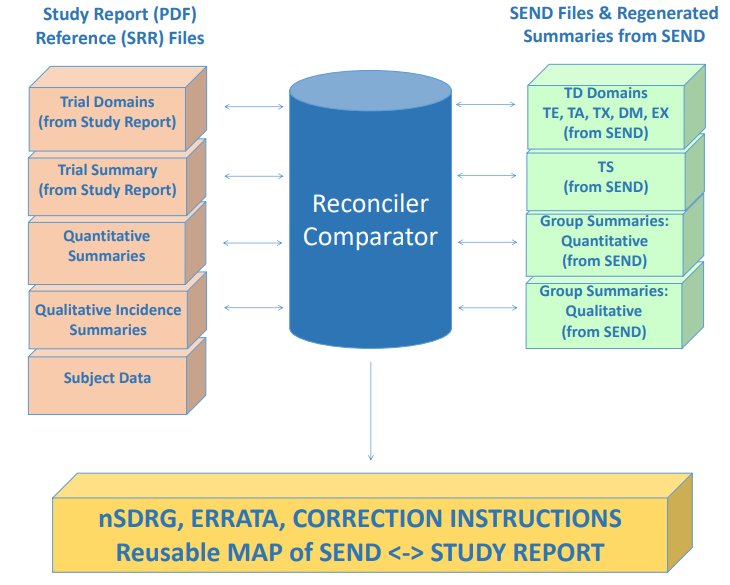
- Summarization, pattern identification and making it available for reconciliation, ready referencing
- Enabling cross study analysis between studies.
- Saving analysis/insights report for presentation, publication, and collaboration
- Biosample availability tracking for search and selection of biosamples for any new assay required and new assay demand generation
- Data tracking and reconciliation of ordered assays on biosamples.
View Poster: Integrating Clinical data for Translational Research