Xbiom support all the workflows, transformations and harmonization required to standardize as-collected data from nonclinical studies or clinical trials. Sourced from LIMS, EDC and Specialty Assay Labs, all data is captured, transformed, and packaged for submission under 21 CFR Part 11 records controls.
Nonclinical Studies and SEND
Nonclinical Studies, started since 2018, are subject to the FDA mandate and are required to be standardized to CDISC SDTM SEND standards and a released version of CDISC Controlled Terminology. Xbiom uses a highly automated, low-touch and no-code process to generate, validate and verify consistency of the SEND data set against its GLP Study Report.
Unlike Clinical Trial data, where the CSR is generated from SDTM standardized as-collected data, in Nonclinical studies the Study Report is generated separately by the Study Director, and the SEND data set is generated by a separate team and necessarily after the analysis that goes into the Study Report is completed. This is because SEND does not have an accompanying ADaM analysis ready data set, but it does include many artifacts that are available only from the analysis done by the Study Director.
The FDA Technical Conformance Guide requires that the SEND data set is capable of generating the same results published in the Study Report. Xbiom includes tools to conduct a complete reconciliation of the SEND data set against the digital tabulation of the reported summary tables in the report. Xbiom includes tools to extract summary tables from Study Reports and the required SEND metadata from the reports. Xbiom also includes all the tools for generating the TS.XPT, the trial design domains (TA, TE, TX, DM), and the Define.XML files. Xbiom also has a built-in eDataValidator for ensuring that the SEND data set meets all the conformance rules including FDA stipulated business rules, and the PMDA rules for Define.XML.
Clinical Study and Submission Data Sets SDTM and ADaM
“As collected” data from patients from the EDC and CRFs, and Biomarker Assay data from bio-banked bio-sample data is ingested through the virtual data room, curated using Xbiom dashboards, and auto-transformed by the Smart-Transformer (ST) into a meta-model (UDM) repository. The entire lifecycle of clinical data through the repository is managed in the Xbiom CDR. The ST uses previously stored mappings from the MDR library, to auto-generate the SDTM with a selected IG and CT. Both the IG and CT are managed in the MDR, which is integrate with its CDR.
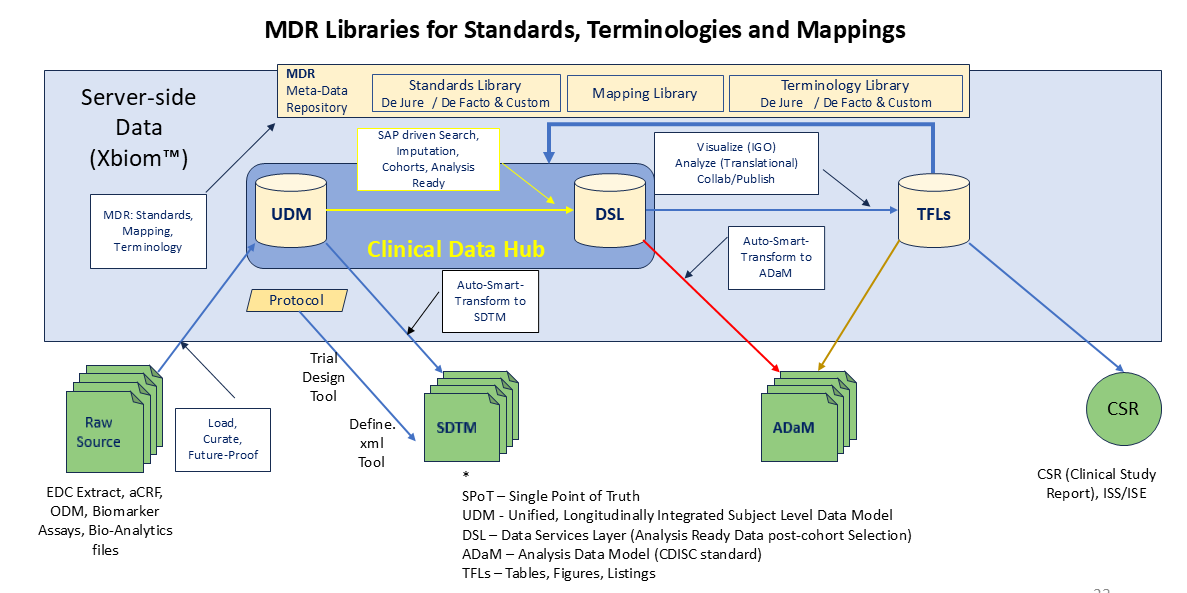
The UDM serves the ST and search query masks with curated As-Collected data so that authorized, qualified bio-statisticians can read, interpret the SAP (Statistical Analysis Plan), and search, find and generate the listings for analysis ready cohorts, and generate the flags needed to generate the ADSL, BDS and OCCDS files of ADaM. The analysis ready data is layer on to the UDM create a Data Services Layer (DSL). The UDM and DSL together form the CDH (Clinical Data Hub) of the CDR. Xbiom’s ST prepares the ADaM data sets either using the previously generated mapping in the MDR, or with programmer generated scripts using R, Python or SAS on the SCE (Statistical Computing Environment).
TFLs may be generated directly using the DSL using the on-board statistical tools and the ability to add custom scripts to generate the required tabulation or figures and charts. It is also possible to generate the TFLs from the ADaM using traditional programming. Tools for generating the Trial Design domains, TS.XPT, and the Define.XML are included.

