Abstract
Large Language Models (LLMs) have the potential to significantly accelerate therapeutics development while reducing costs and increasing innovation in the biopharmaceutical industry. However, two critical barriers prevent widespread adoption of this technology.
First, competitive sensitivities and data sovereignty requirements mandate that clinical trial and nonclinical study data remain within pharmaceutical companies’ secure networks. This necessitates in-house LLM deployment to prevent potential data leakage throughout the development lifecycle – from early-stage study design and data standardization to late-stage regulatory analysis and report generation.
Second, the complexity of existing data management systems creates significant user adoption barriers. These sophisticated platforms require extensive training and deep technical familiarity, leading to user reluctance and limited engagement among the scientists and managers who would benefit most from these tools.
This paper presents a comprehensive Retrieval Augmented Generation (RAG) framework that addresses both challenges by securely integrating LLM capabilities with the Xbiom™ unified data repository. The framework combines the reasoning and generative capabilities of LLMs with Xbiom’s built-in retrieval, analysis, and visualization functions across diverse study types and data formats.
The RAG-enabled system transforms the user experience from complex application navigation to intuitive chat-based interactions. These conversations trigger specialized agents that retrieve relevant information for data management, standardization, analysis, and reporting tasks. Results are delivered either as generated text summaries or direct links to specific Xbiom applications.
By eliminating the complexity barrier, this approach enables mid-level and senior-level users to directly engage with study data for insight generation and decision-making. The automation of routine activities – including data discovery, retrieval, and reporting – reduces development lifecycle time and costs by minimizing manual data wrangling and low-level programming requirements.
Barriers to automating clinical data management in BioPharma
BioPharma companies and regulatory agencies accumulate vast study data across therapeutic programs, but it remains siloed by department in separate systems. Accessing, analyzing, or extracting insights requires specialists trained on separate systems:
- Document management for regulatory files
- Data lakes for raw lab data
- Metadata repositories for standards and governance
- Configuration management for data files and SAS/R programs
- Statistical computing environments for analysis plan execution
- Document systems for research and visualizations
If these systems are implemented in separate applications that do not have tight integration or have weak inter-operability that presents a very tough environment that makes adoption very difficult. Each system requires extensive training. Strategic decision makers and managers cannot easily access data or insights worked on by multiple specialists. Cross-study analysis is nearly impossible despite major digitization investments in standardization and harmonization for the purposes of exchange; but not for future-proofed repositories that can hold all studies – irrespective of their type or age.
Some of these challenges can be overcome by an integrated environment or with the use of a integrated solutions platform with a unified repository. The unified repository ensures data integration. The solution platform ensures that the applications interoperate, and data matured in one facility is available to the other facilities based on the user roles and authorization. The unified repository is important for a future-proofed way to hold and make available disparate study data and insights. Departmental barriers will still exist but they can be overcome with corporate commitment.
Xbiom – Integrated Solution Platform with a Unified Study Data Repository
The Xbiom platform centers on a unified repository holding all structured, unstructured, and semi-structured data from early to late-stage development. It functions as both a clinical data management system and safety hub, containing as-collected data, standardization metadata, analysis metadata, and annotated results.
Integrated sub-systems that surround the Xbiom repository and platform include:
- Data exchange portal for inter-organizational sharing (VDR)
- Self-organizing document management (Orchestrating DMS)
- ML-enabled smart transformation (ST)
- Standards and terminology reference library & governance (MDR)
- Indexed search across unified data and documents (UDM)
- LLM extraction of instructions from protocols and reports (Text2Param)
- Analysis metadata driven by statistical plans (Insights)
- Automated standardized dataset generation (SDTM)
- Validators and Reconcilers (eDV)
- Analysis and visualization for templated or ad hoc scripts (IGO)
- Automated annotated results generation with insight storage (aTFL)
Users interact with Xbiom based on their job function and access the applications they need. They need to know the features and functions of the applications, and how they are integrated. Xbiom returns information based on the user’s request, and the nature of the data – as datasets, listings, dashboards with metadata, filterable displays of categorical or multi-dimensional data, charts, tabulations, results of analysis such as end-points, statistical trends and p-values.
Barriers to User adoption
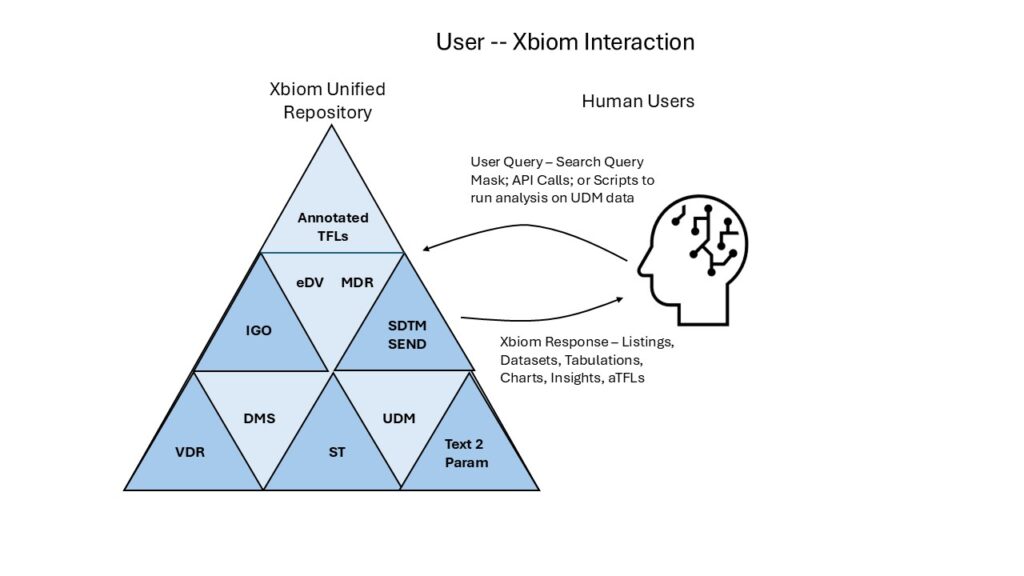
Each facility in Xbiom handles data management, transformation, reporting, and analysis differently based on the data types and the functional needs. Separate departments use them with minimal overlap. Each system has specialized features and displays requiring extensive training and regular use to maintain proficiency. Operations staff who wrangle data and do procedural tasks will be proficient users and they can work effectively within their functional areas. Tactical decision makers who need to slice and dice the data and prepare story telling presentations with tabulations and charts, or who need to coordinate and supervise cross-functionally, rarely gain proficiency and rely on support staff.
Strategic decision makers rarely use these systems enough to stay proficient, so they rely on assistants to retrieve and present data. This creates problems: requests are often nuanced and get misunderstood, leading to multiple rounds of searching and analysis. Complete cycles can take days or weeks.
Navigating complex menus, learning such specialized software, or breaking tasks into rigid steps, cause friction in the User’s need to cycle between questions and answers. Without direct access to data, decision makers lose the intimacy with answers they need for quicker, effective strategic and tactical decisions. The barriers are:
- Complex user interfaces and controls to access necessary features in every functional module
- Precise, correct queries to the application will return precisely correct responses from the stored data and metadata. But nuanced queries of human users return unexpected responses triggering follow on queries
- Different users like to see the same data in different formats – some prefer data in listings, or organized data sets, other prefer tabulation models or charts. Learning and using the controls to get results in their preferred format is time consuming, and distracting.
- Better User experience if a summarized response is available with the ability to drill down to the supporting evidence of data and charts
A natural conversational interface to improve adoption
A simple chat interface to allow users to talk to such specialized software breaks down the barriers faced by most users. It lets them go between their questions to answers in seconds. There are no menus to navigate and select. Nuanced or unclear questions are clarified and leads the user towards more precise question that get them to their answer.
The simple search chat box of the past decades has now become mainstream in all the prominent LLMs. Inserting an LLM in between the User and the specialized application in Xbiom transforms the user experience to one of discovery, search, inference and testing their reasoning for solving problems.
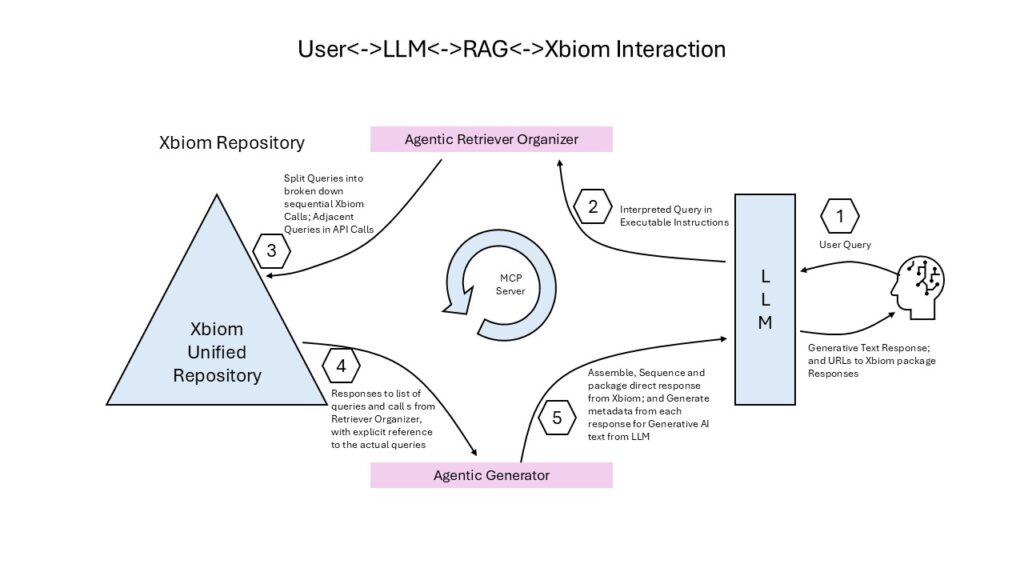
The LLM interprets the query, converting it to the API calls understood by Xbiom, using the RAG (Retrieval Augmented Generation) process. Disambiguation of the query, clarifying challenges, spawning agents that get expected, and anticipated, results from the Xbiom applications speeding up the process of getting answers. The LLM helps in generating readable text that summarizes or explains the content in the answers retrieved from the applications in Xbiom.
LLM + RAG + Xbiom is a perfect match for intelligent automation
LLMs, RAG, and Xbiom each have their limitations and their strengths, and each of their strengths overcome the limitations of the other two.
LLMs Can:
- Answer question posed in English
- Edit questions for clarity and context
- Summarize document and extract parameters
- Generative text from retrieved responses
- Explain complex responses
- Generate scripts and code
LLMs Cannot:
- Browse or get answers from applications or intra-net sources
- Remember conversations outside of the session
- Perform calculations reliably
- Access data repositories to get data or metadata
- Take actions or perform workflows
But RAG Can:
- Connect LLM responses to the current, specific, proprietary information in a Xbiom repository
- Access real-time data, metadata, unstructured documents from specialized repositories like Xbiom
- Carry source attribution, provenance, and supporting documentary or data evidence
- Work with constantly changing data, such as accumulated studies in the repository
- Customize and condition responses based on user roles and privileges
- Reduce hallucinations by anchoring responses to retrieved content
RAG Cannot:
- Multi-step logic and synthesizing multiple data/document sources
- Ensuring consistency between data sources
- Perform rigorous analytical computations on retrievable data
- Fill gaps in the data response
- Adapt retrieval strategies based on user behavior
- Semantic limitation – disambiguating queries
Xbiom Can:
- Synthesize information from multiple data/document sources
- Maintains internal consistency and rigor in its data and information responses
- Perform rigorous analytical computation on its data
- Fill gaps in data response, and LLM can provide contextual feedback for user
- Adapt, remember user behavior and drive RAG to generate preferred responses to users
- Use its MDR to disambiguate queries and semantically consistent responses
The right LLM for Pharma Therapeutic Development Use
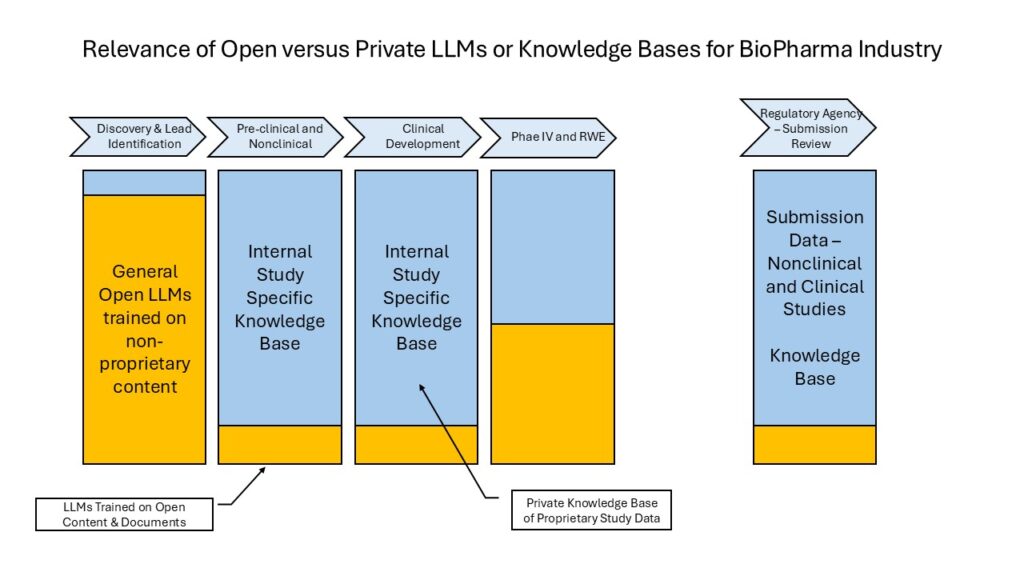
Open LLMs like ChatGPT, Gemini, LLAMA, Claude are trained on vast amounts of books, papers, articles and websites. They have a very large footprint and they are delivered as a solution on a public cloud. These cannot be used by BioPharma enterprises for any of their nonclinical or clinical development phases because any integration will expose their proprietary study data to these LLM systems through the queries sent to them by users. That would be an unacceptable breach of data security.
Narrower, biomedically trained LLMs such as BioGPT, PubMedGPT, MedAlpaca, TxGemma, GatorTron, and Med-PaLM2 are also trained on openly available content including papers, articles, public registries, and websites. Even generally trained LLMs with smaller footprints are available (Gemma, Llama, DeepSeek) and they may be used as well. Their smaller footprint allows easy installation within the firewalls of a BioPharma enterprise, making them suitable for integration with a RAG and for use with a data and knowledge base such as Xbiom.
Xbiom is a big-data, indexed repository with a unified data model for holding any, and all, studies or clinical trials that may have been conducted by a Pharma over the years. It has a versatile and robust search engine to return complex structured queries as well search within documents. It is embedded with applications for transformation, standardization, analysis, visualization and generation of annotated TFL objects that carry insights of researchers.
LLMs are static in that they do not gain new information unless they are re-trained at great expense and effort. The enterprise indexed repository for studies and trials, on the other hand is dynamic and constantly growing as new studies are started and completed.
RAG, or Retrieval Augmented Generator, is an effective way to extend the frozen “memory” of a trained LLM. It is an active bridge between the text capable intelligent LLM and the data intensive unified store that can return request for data, metadata, insights and a combination of these.
RAG is an innovative framework that enhances large language model outputs by incorporating external, dynamically retrieved data. The system essentially consists of two major components: a retriever, which searches a curated database or unified repository for relevant data, information and documents based on the input query; and a generator, which conditions its response on both the user prompt and the retrieved context. This combination aims to ground the answer in up-to-date, domain-specific information that might not be fully captured in the model’s original training.
Xbiom RAG System: Data-Information-Knowledge Framework
Xbiom’s RAG system uses retrieval and generation functions to access study data, metadata, and stored insights. These functions support three levels of data interaction: procedural (curation and standardization to SDTM/SEND), tactical (data preparation and viewing), and strategic (scientific analysis and insight development).
Integrating LLM with RAG automates these functions with enhanced depth and rigor. The procedural, tactical, and strategic aspects determine what information is retrieved and how it should be reported. Agents between the LLM and Xbiom handle the complete workflow from user query to search execution to response delivery whether directly to users or through AI-generated text responses.

In some cases the response is directly returned to the user, while in some cases the retrieved response may be further conditioned by the LLM using Generative AI text or other techniques including tabulation and figures to give the user a more meaningful response. The figure below shows how the Human User Interacts with the Xbiom repository and tools through the LLM and the RAG structures.
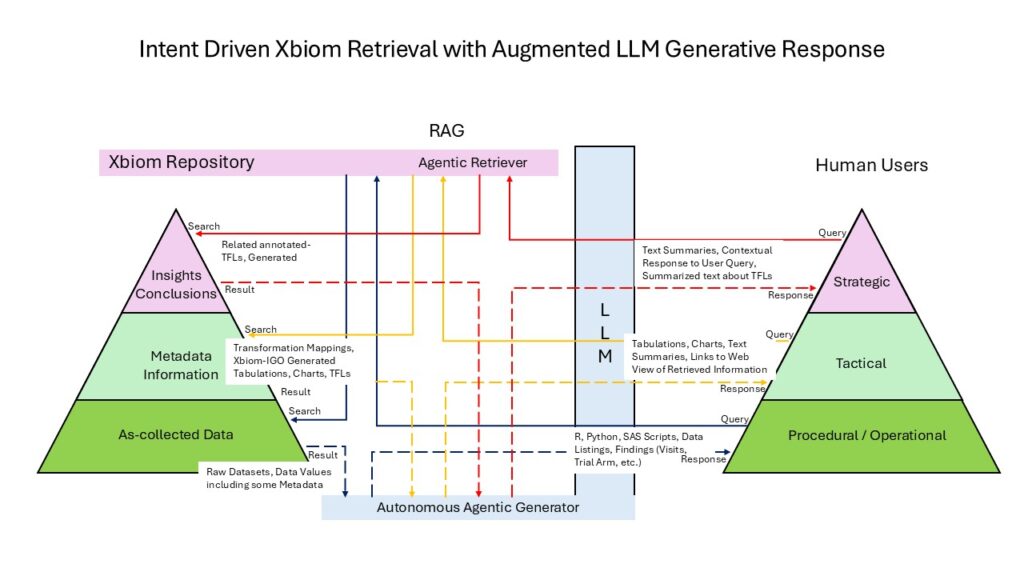
Xbiom’s Three-Layer Architecture organizes content across three maturity levels that align with different user inquiry types:
- Data Layer: As-collected study data (21 CFR Part 11 compliant) from in-vivo studies and in-vitro assays
- Information Layer: Metadata, standardized formats, analysis-ready datasets, and visualizations
- Knowledge Layer: Insights, TFLs (Tables, Figures, Listings), and study reports
Contact [email protected] or visit our website at www.pointcross.com
PointCross Inc.
Foster City, CA 94404
Tel: +1 (844) 382 – 7257

